Research Article - Onkologia i Radioterapia ( 2022) Volume 16, Issue 9
Hybrid deep CNN-LSTM network for breast histopathological image classification
Ankita Patra1, Santi Kumari Behera2 and Nalini Kanta Barpanda12Department of Computer Science and Engineering, VSSUT, Odisha, India
Received: 08-Sep-2022, Manuscript No. OAR-22-74101; Accepted: 28-Sep-2022, Pre QC No. OAR-22-74101 (PQ); Editor assigned: 10-Sep-2022, Pre QC No. OAR-22-74101 (PQ); Reviewed: 24-Sep-2022, QC No. OAR-22-74101 (Q); Revised: 26-Sep-2022, Manuscript No. OAR-22-74101 (R); Published: 29-Sep-2022
Abstract
Breast cancer is a serious public health concern since it is associated with the highest rates of cancer-related morbidity globally. The likelihood of effective treatment and deaths are increased with early detection. Sadly, it is a challenging and urgent task requiring pathologists' skills. Automatic breast cancer identification based on histopathological pictures dramatically benefits patients and their future. In order to categorize breast histopathology images, this research intends to offer a deep learning technique that combines a Convolutional Neural Network (CNN) with Long Short-Term Memory (LSTM). In this system, deep feature extraction is performed by CNN, while detection utilizing the extracted feature is performed by LSTM. For experimentation, BACH 2018 dataset is used, which includes 800 images of four kinds, i.e., normal, benign, invasive, and in situ. The achieved accuracy, sensitivity, specificity, precision, and computational time are 98.1%, 95.5%, 96.4%, 97.2%, and 24 seconds respectively
Keywords
deep learning, breast cancer, GLCM, HOG, LSTM
Introduction
The greatest cause of death for women globally between the ages of 20 and 59 is breast cancer. However, up to 80%, more people can survive breast cancer if diagnosed early [1]. To understand various bioactivities, the analysis of microscopic images of different human organs and tissues is crucial in biomedicine. One of the most significant jobs in the microscopical study is classifying images (tissues, organs, etc.). Based on the classification of microscopic images, numerous applications have been developed
CNN's need to be trained on high-resolution images directly, which requires a strong linear transformation at the input layer. This is so they can find oddities in medical images. In this integrated technique for medical abnormality detection using deep CNN, pre-trained deep CNNs are carefully tuned on input images focusing on medical abnormalities before being connected with a class basis function to make abnormality detectors. An alternative to the manual method, a DL classifier was tested on a single-class Histopathology dataset with an accuracy of 92.53% [2]. “Ahmad et al. suggested using DL and transfer learning to classify histopathological images for diagnosing breast cancer. This study used a patch selection approach to classify breast histopathological images based on a small number of training images and transfer learning without losing performance. At first, patches from Whole Slide Images are taken out and fed into the CNN so that features can be taken out. Based on these features, the discriminative patches are chosen and then fed to an Efficient-Net architecture that has already been trained on the ImageNet dataset. An SVM classifier can also be trained using features from an Efficient-Net architecture” [3]. “Sharma et al. showed that, unlike handcrafted approaches, the pre-trained Xception model could classify breast cancer histopathological images based on their size. With an accuracy of 96.25%, 96.25%, 95.74%, and 94.11% for 40×, 100×, 200×, and 400× levels of magnification, respectively, the Xception model and SVM classifier with the radial basis function kernel have given the best and most consistent results” [4]. Zou et al. create a new Attention High-order Deep Network (AHoNet) by putting a high-order statistical representation and an attention mechanism into a residual convolutional network at the same time [5]. “AHoNet first uses an efficient channel attention module with non-dimensionality reduction and local cross-channel interaction to get local salient deep features of breast cancer pathological images. Then, their second-order
covariance statistics are estimated further through matrix power normalization. This gives a more stable global feature presentation of breast cancer pathological images. We use the public BreakHis and BACH breast cancer pathology datasets to test AHoNet in depth. Experiments show that AHoNet gets the best patient-level classification accuracy of 99.29% on the BreakHis database and 85% on the BACH database” [5]. “Hao et al. recognized breast cancer histopathological images using deep semantic features and Grey Level Co-Occurrence Matrix (GLCM) features. Using the pre-trained DenseNet201 as the basic model, part of the convolutional layer features of the last dense block are extracted as the deep semantic features. These are combined with the three-channel GLCM features, and the Support Vector Machine (SVM) is used to classify. For 40, 100, 200, and 400, the highest image-level recognition accuracy is 96.75%, 95.21%, 96.57%, and 93.15%, respectively” [6]. Kashyap et al. developed a model for classifying breast cancer histopathology images based on DL [7]. They called it the Stochastic Dilated Residual Ghost (SDRG). Overall, the SDRG model was 95.65% accurate at putting images into categories and 99.17% accurate when it came to being precise. Ding et al. suggest a method for classifying breast histopathology images that combine features from different levels [8]. First, the attention mechanism and convolutions combine shallow and deep semantic features. Then, a new weighted cross entropy loss function is used to deal with false negatives and false positives. And finally, the wrong estimates of some patches are fixed by correlating information about where they are. The proposed method has a 99.0% accuracy and a 99.9% AUC.
Material and Methodology
This section detailed the dataset and proposed methodology.
About Dataset
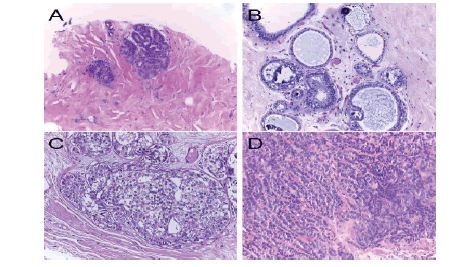
The Breast Cancer Histology Challenge (BACH) 2018 dataset contains all slide images from breast histology microscopes stained with Hematoxylin and Eosin (H&E). A total of 800 microscope pictures make up the collection, with 200 each representing normal, benign, in situ, and invasive carcinomas. The .tiff format, RGB colour model, 2048×1536-pixel size, 0.42 m×0.42 m pixel scale, and image-wise labelling are all characteristics of microscopy images. Figure 1 illustrates the sample photographs.
Figure 1:. BACH 2018 dataset. (a) In situ carcinoma, (b) invasive carcinoma, (c) Normal tissue(d) Benign Methodology
METHODOLOGY
The proposed architecture was made by combining a CNN and a LSTM network, which are briefly explained below. Our proposed hybrid CNN-LSTM- DL model comprises two important parts. The first part includes layers called "pooling" and "convolutional." In these layers, complex scientific operations have been done to build the characteristics of the input data. The second part is made up of LSTM and dense layers to use the features that were generated. The convolutional layer comprises a group of kernels used to figure out a tensor of feature maps [9]. Using "stride(s)," these kernels change the shape of the whole input so that the dimensions of the output volume are integers [10]. After the convolutional layer is used to run the striding process, the size of an input volume goes down. So, zero padding is needed to fill in an input volume with zeros and keep its size when using low-level features [11]. The pooling layer takes a smaller sample of an input dimension to cut down on the number of parameters [12]. The most common method is max pooling, which takes an input region and finds its highest value. The FC layer is a classifier that uses features from the convolutional and pooling layers to make a decision [13].
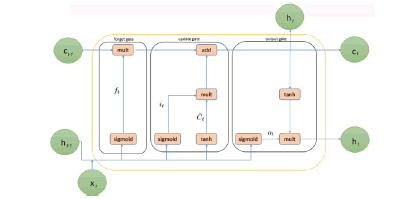
Recurrent Neural Networks (RNNs) are made better by longterm, short-term memory. To solve the vanishing and exploding gradient problem, LSTM proposes memory blocks instead of the usual RNN units [14]. The main difference between it and RNNs is that it adds a cell state to save long-term states. An LSTM network can remember information from the past and link it to information from the present [15]. LSTM is made up of three gates, such as an input gate, a "forget" gate, and an output gate, where it is the current input, Ct and Ct 1 are the current and previous cell states, and ht and ht 1 are the current and prior outputs. Figure 2 shows how an LSTM works on the inside
Figure 2: The internal structure of LSTM
In this study, a combined method was made to automatically classify the histopathological images of the breast. This architecture's structure was made by putting together CNN and LSTM networks. CNN pulls out complex features from images, and LSTM is used as a classifier
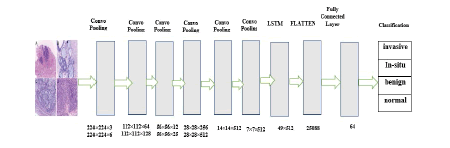
Figure 3 shows the proposed hybrid network for the classification of histopathological images. The network comprises 20 layers: 12 convolutional layers, five pooling layers, one FC layer, one LSTM layer, and one soft max output layer. Each convolution block combines two or three 2D CNNs, one pooling layer, and a dropout layer with a 25% dropout rate. The ReLU function turns on the convolutional layer, which has 3×3 kernels and is used to pull out features. The max-pooling layer, which has a size of 22 kernels, is used to make an image smaller. In the last part of the architecture, time information is taken from the function map by moving it to the LSTM layer. The output shape is found after the convolutional block (none, 7, 7, 512). By using the reshape method, the LSTM layer's input size has grown (49, 512). After looking at the features, the architecture sorts the histopathological images through a fully connected layer to predict which categories they belong to (benign, invasive, insitu, and normal).
Figure 3: Proposed hybrid deep CNN-LSTM network for breast histopathological image classification
Result and Discussion
The CNN and CNN-LSTM networks were built on an Intel(R) Core (TM) i7-12th generation processor using MATLAB 2021a. A Graphical Processing Unit (GPU) NVIDIA RTX 3050 Ti with 4 GB and 16 GB RAM was also used to run the experiments. Experiments have shown that the maximum number of epochs in the proposed network is 125. The learning rate is 0.0001, and the number of convolutional layers is 12. The research framework was put into place and tested using 800 images with 200 samples in each category. The 5-fold crossvalidation technique was used to get the results. Table 1 shows the results of running the proposed method and other classifiers on the same data set.
Tab.1. Performance of classifiers for breast histopathological image classification
| Classifier | SVM | KNN | ANN | Proposed CNN-LSTM |
|---|---|---|---|---|
| Accuracy (%) | 94.7 | 91 | 92.7 | 98.1 |
| Sensitivity (%) | 96.6 | 94.2 | 92.3 | 95.5 |
| Specificity (%) | 93.4 | 82.5 | 93.5 | 96.4 |
| Precision (%) | 91.3 | 84.2 | 84.7 | 97.2 |
| Time (second) | 32 | 25 | 36 | 24 |
From Table 1, the classification with the Proposed DL Classifier with CNN-LSTM has a 98.1% overall accuracy, which is higher than those obtained using other methods. According to Table 1, the image's True Positive values should always be greater for better results, and the False Negative should be as low as possible. The experiment results show that when employing a Proposed DL Classifier with DNN-LSTM, the overall accuracy is higher than standard classifiers. Based on tests on various photographs, the proposed method appears to work well in both circumstances when the elements in the image are unclear and distinguishable from the background.
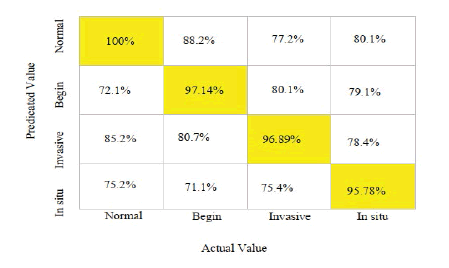
From Table 2, the Proposed DL Classifier with CNN-LSTM is used to classify the data, which analyses the characteristics that belong to the Predicted output and the Target Class. The Machine Learning Classifier has a 100% accuracy rate for normal, 97.14% in the beginning, 96.89% for invasive, and 95.78% in situ, as illustrated in Figure 4.
Tab.2. Comparison of classifier values concerning the confusion matrix
| Classifier | TP(%) | FP(%) | FN(%) | TN(%) |
|---|---|---|---|---|
| SVM | 42.8 (%) | 56.6 (%) | 43.4(%) | 58 (%) |
| KNN | 44.5 (%) | 55.2 (%) | 42.8(%) | 54.5 (%) |
| ANN | 45.8 (%) | 57.5 (%) | 44.2(%) | 55.2 (%) |
| Proposed CNN-LSTM | 48.5 (%) | 64.9 (%) | 35.1(%) | 51.5 (%) |
Figure 4: Confusion matrix of hybrid CNN-LSTM for breast histopathological image classification
Acknowledgement
The authors are thankful to Faculty Research Grant, SRIC, Sambalpur University, for providing financial assistance for conducting this research.
References
- Wei L, Yang Y, Nishikawa R M. Micro calcification classification assisted by content-based image retrieval for breast cancer diagnosis. Pattern Recognit. 2009; 42:1126–1132.
- Carvalho ED, Antonio Filho OC, Silva RR, Araujo FH, et al. Breast cancer diagnosis from histopathological images using textural features and CBIR. Artif Intell Med. 2020; 105:101845.
- Ahmad N, Asghar S, Gillani SA. Transfer learning-assisted multi-resolution breast cancer histopathological images classification. Vis. Comput. 2022;38:2751-2770.
- Sharma S, Kumar S. The Xception model: A potential feature extractor in breast cancer histology images classification. ICT Express. 2022;8:101-8.
- Zou Y, Zhang J, Huang S, Liu B. Breast cancer histopathological image classification using attention highâ?order deep network. Int J Imaging Syst. Technol. 2022; 32:266-279.
- Hao Y, Zhang L, Qiao S, Bai Y, Cheng R, et al. Breast cancer histopathological images classification based on deep semantic features and grey level co-occurrence matrix. Plos one. 202217:e0267955.
- Kashyap R. Breast cancer histopathological image classification using stochastic dilated residual ghost model. Int J Inf Retr Res. (IJIRR). 2022; 12:1-24.
- Ding WL, Zhu XJ, Zheng K, Liu JL, You QH. A multi-level feature-fusion-based approach to breast histopathological image classification. Biomed Phys Eng Express. 2022;8:055002.
- Hasan AM, Jalab HA, Meziane F, Kahtan H, Al-Ahmad AS. Combining deep and handcrafted image features for MRI brain scan classification. IEEE Access.2019;7:79959-79967.
- Gu J, Wang Z, Kuen J, Ma L, Shahroudy A, et al. Recent advances in convolutional neural networks. Pattern recognit. 2018;77:354-377.
- Kutlu H, Avcı E. A novel method for classifying liver and brain tumours using convolutional neural networks, discrete wavelet transform and long short-term memory networks. Sensors. 2019; 19:1992.
- Lundervold AS, Lundervold A. An overview of deep learning in medical imaging focusing on MRI. Z Med Phys.2019;29:102–127.
- Chang P, Grinband J, Weinberg BD, Bardis M, Khy M, et al. Deep-learning convolutional neural networks accurately classify genetic mutations in gliomas. Am J Neuroradiol. 2018; 39:1201–1207.
[CrossRef]
- Hochreiter S. The vanishing gradient problem during learning recurrent neural nets and problem solutions. Int J Uncertain Fuzziness Knowl-Based Syst. 1998;6:107-116.
- Chen G. A gentle tutorial of recurrent neural network with error backpropagation. arXiv Prepr. 2016.