Research Article - Onkologia i Radioterapia ( 2022) Volume 16, Issue 7
Brain tumour segmentation using SRGB colour space-based density assessment
Nibedita Pati1*, Millee Panigrahi2 and Krishna Chandra Patra32Department of Electronics and Telecommunication Engg,Trident Academy of Technology, Bhubaneswar , Od, Iceland
3Department of Electronics,Sambalpur University,Odisha - 768019, India
Nibedita Pati, Department of Electronics, Sambalpur University Institute of Information Technology, Odisha - 768019, India, Email: nibedita.tech2007@gmail.com
Received: 16-Jul-2022, Manuscript No. OAR-22-69374; Accepted: 31-Jul-2022, Pre QC No. OAR-22-69374 (PQ); Editor assigned: 18-Jul-2022, Pre QC No. OAR-22-69374 (PQ); Reviewed: 22-Jul-2022, QC No. OAR-22-69374 (Q); Revised: 24-Jul-2022, Manuscript No. OAR-22-69374 (R); Published: 01-Aug-2022, DOI: https://doi.org/10.5281/zenodo.8115477
Abstract
Medical image processing helps diagnose diseases early. Brain tumour segmentation is a medical imaging speciality. Computer vision and machine learning help doctors diagnose diseases effectively. This study uses Standard RGB (SRGB) density analysis to isolate brain tumours on MRI images. Input intensity values are normalized using SRGB colour space and a Gaussian filter to identify tumours from the background. Adaptive threshold identifies brain MRI tumour spaces. Brain tumour space is derived using area and density functions. Applying morphological functions eliminates false positives to detect the accurate tumour space. The proposed technique is evaluated using recall, precision, and F–measure.
https://www.mobafire.com/profile/marmaristeknekirala12-1120487?profilepage http://www.effecthub.com/user/3725743 http://hawkee.com/profile/4949273/ https://www.youmagine.com/marmaristeknekirala12/designs https://www.myminifactory.com/users/Marmaristeknekirala12 http://onlineboxing.net/jforum/user/editDone/250019.page http://qooh.me/Marmaristekne https://linktr.ee/marmaristeknekirala12 https://pubhtml5.com/homepage/rskea/ https://telegra.ph/Marmaristeknekirala12-09-05 https://www.diggerslist.com/64f721ff718d5/about https://allmyfaves.com/Marmaristeknekirala12 https://www.metal-archives.com/users/Marmaristeknekirala12 https://www.catchafire.org/profiles/2490257/ https://www.fimfiction.net/user/633629/Marmaristeknekirala12 https://www.hebergementweb.org/members/marmaristeknekirala12.548722/ https://www.sqlservercentral.com/forums/user/marmaristeknekirala12 https://www.twitch.tv/marmaristeknekirala12/about https://www.roleplaygateway.com/member/Marmaristeknekirala12/ https://ko-fi.com/marmaristeknekirala12#paypalModal https://www.provenexpert.com/marmaristeknekirala12/ https://www.intensedebate.com/people/marmaristekne https://www.indiegogo.com/individuals/35150686 https://visual.ly/users/marmaristeknekirala/portfolio https://slides.com/marmaristeknekirala12 https://letterboxd.com/marmaristekne/ https://micro.blog/Marmaristeknekirala https://fliphtml5.com/dashboard/public-profile/pudlf https://community.windy.com/user/marmaristekne https://speakerdeck.com/marmaristeknekirala12 https://trello.com/u/marmaristeknekirala/activity https://www.redbubble.com/people/Marmaristekne/shop?asc=u https://myanimelist.net/profile/Marmaristekne https://3dwarehouse.sketchup.com/user/12639f7f-92aa-411b-a067-e7deea565fbf/Marmaristeknekirala12-M https://www.wattpad.com/user/Marmaristekne https://www.goodreads.com/user/show/169647299-marmaristeknekirala12 https://sourceforge.net/u/marmaristekne/profile https://tr.gravatar.com/marmaristeknekirala https://tr.pinterest.com/marmaristeknekirala/ https://www.mapleprimes.com/users/Marmaristeknekirala12 https://medium.com/@marmaristeknekirala https://www.ted.com/profiles/44959924 https://www.infragistics.com/community/members/d358780bec8a5f20d6f4563b7fd9e569ff4f3a39?_ga=2.74596625.2087551855.1693994141-821832157.1693994141 https://www.metooo.io/u/64f84eca6d5e6b4d2dc9d465 https://app.roll20.net/users/12383781/marmaristeknekirala12-m https://list.ly/marmaristeknekirala/newsfeed https://giphy.com/channel/Marmaristeknekirala12 https://www.tumblr.com/marmaristeknekirala12 https://dzone.com/users/4989214/marmaristeknekirala12.html https://www.creativelive.com/student/marmaristeknekirala12-marmaristeknekirala12?via=accounts-freeform_2 https://gab.com/Marmaristeknekirala12 https://sketchfab.com/Marmaristeknekirala12 https://www.flickr.com/people/httpsmarmaristeknekiralacomtr/ https://hub.docker.com/u/marmaristeknekirala12 https://forums.autodesk.com/t5/user/viewprofilepage/user/self https://mastodon.online/@Marmaristeknekirala12 https://wefunder.com/marmaristeknekirala12 https://calis.delfi.lv/profils/ https://os.mbed.com/users/marmaristeknekirala1/ https://notionpress.com/author/921940# https://guides.co/a/marmaristeknekirala12-marmaristeknekirala12/ https://www.bahamaslocal.com/userprofile/1/239817/Marmaristeknekirala12.html http://molbiol.ru/forums/index.php?showuser=1296710 https://www.credly.com/users/marmaristeknekirala12-marmaristeknekirala12/badges https://www.facer.io/user/irCYaMEYX0 https://www.mobafire.com/profile/marmarishome12-1119055?profilepage https://www.wishlistr.com/profile/ http://www.effecthub.com/user/3724210 http://hawkee.com/profile/4884005/ https://www.youmagine.com/marmarishome/designs https://www.myminifactory.com/users/marmarishome12 http://onlineboxing.net/jforum/user/edit/248549.page http://qooh.me/account/home/ https://band.us/band/92152039 https://linktr.ee/marmarishome12 https://pubhtml5.com/homepage/iprhm/ https://telegra.ph/marmarishome12-08-24 https://www.diggerslist.com/64e719eb6d769/about https://www.facer.io/user/WJmAjPpNr9 https://allmyfaves.com/marmarishome12 https://www.metal-archives.com/users/marmarishome12 https://www.catchafire.org/profiles/2479894/ https://www.fimfiction.net/user/630254/marmarishome12 https://www.hebergementweb.org/members/marmarishome12.542713/ https://www.sqlservercentral.com/forums/user/marmarishome12 https://www.twitch.tv/marmarishome12/about https://www.roleplaygateway.com/member/marmarishome12/ https://ko-fi.com/marmarishome12 https://www.provenexpert.com/marmarishome12/ https://www.intensedebate.com/people/marmarishome12 https://www.indiegogo.com/individuals/35056645 https://visual.ly/users/marmarishome/portfolio https://slides.com/marmarishome12 https://letterboxd.com/marmarishome12/ https://micro.blog/marmarishome12 https://fliphtml5.com/dashboard/publications https://community.windy.com/user/marmarishome12 https://www.longisland.com/profile/marmarishome12 https://speakerdeck.com/marmarishome12 https://trello.com/u/marmarishome/activity https://www.redbubble.com/people/marmarishome12 https://disqus.com/by/marmarishome12/about/ https://myanimelist.net/profile/marmarishome12 https://3dwarehouse.sketchup.com/user/c6ed99ef-400c-4a3d-af93-11bfcb2be913/marmarishome12-M https://www.wattpad.com/user/marmarishome12 https://www.goodreads.com/user/show/169358554-marmarishome12 https://sourceforge.net/u/marmarishome12/profile https://en.gravatar.com/marmarishomee4e8b75111 https://tr.pinterest.com/marmarishome/ https://www.mapleprimes.com/users/marmarishome12 https://medium.com/@marmarishome https://www.ted.com/profiles/44878861 https://www.infragistics.com/community/members/5d36dfb4d9fe1a193a81f1559de3c43d20e17704 https://marmarishome12.livejournal.com/profile https://www.metooo.io/u/64ec674f6d5e6b4d2dc02b90 https://hubpages.com/@marmarishome12 https://app.roll20.net/users/12349873/marmarishome12-m https://list.ly/marmarishome/ https://giphy.com/channel/marmarishome12 https://www.tumblr.com/blog/marmarishome https://dzone.com/users/4984261/marmarishome12.html https://www.creativelive.com/student/marmarishome12-marmarishome12?via=accounts-freeform_2 https://gab.com/marmarishome12 https://sketchfab.com/marmarishome12 https://www.flickr.com/people/199033566@N03/ https://devpost.com/marmarishome https://hub.docker.com/u/marmarishome12 https://forums.autodesk.com/t5/user/viewprofilepage/user/self https://profiles.wordpress.org/marmarishome12/ https://mastodon.online/@marmarishome12 https://wefunder.com/marmarishome https://seedandspark.com/user/marmarishome12 https://calis.delfi.lv/profils/lietotajs/311777-marmarishome12/ https://os.mbed.com/users/marmarishome12/ https://notionpress.com/author/918713# https://my.desktopnexus.com/marmarishome12/#ProfileComments https://guides.co/a/marmarishome12-marmarishome12/ https://www.bahamaslocal.com/userprofile/1/237330/marmarishome12.html http://molbiol.ru/forums/index.php?showuser=1295383 https://www.credly.com/users/marmarishome12-marmarishome12/badges https://orcid.org/0009-0009-4483-3745 https://www.mobafire.com/profile/marmarisboatcharter12-1124453?profilepage https://www.youmagine.com/marmarisboatcharter12/designs https://www.myminifactory.com/users/Marmarisboatcharter12 http://qooh.me/MarmarisSS https://linktr.ee/marmarisboatcharter12 https://pubhtml5.com/homepage/pdrml/ https://telegra.ph/Marmarisboatcharter12-10-06 https://www.diggerslist.com/651fbb00ae0f3/about https://allmyfaves.com/Marmarisboatcharter12 https://www.metal-archives.com/users/Marmarisboatcharter12 https://www.catchafire.org/profiles/2517163/ https://www.fimfiction.net/user/643467/Marmarisboatcharter12 https://www.hebergementweb.org/members/marmarisboatcharter12.562475/ https://www.sqlservercentral.com/forums/user/marmarisboatcharter12 https://www.twitch.tv/marmarisboatcharter12/about https://www.roleplaygateway.com/member/Marmarisboatcharter12/ https://www.provenexpert.com/marmarisboatcharter12/?mode=preview https://www.intensedebate.com/people/Marmarisboat https://www.indiegogo.com/individuals/35445398 https://visual.ly/users/marmarisboatcharter/portfolio https://slides.com/marmarisboatcharter12 https://letterboxd.com/marmarisboatcha/ https://fliphtml5.com/dashboard/public-profile/sfskb https://community.windy.com/user/marmarisboat https://speakerdeck.com/marmarisboatcharter12 https://trello.com/u/marmarisboatcharter https://myanimelist.net/profile/Besthairtranss https://3dwarehouse.sketchup.com/user/5acb3c60-4aca-43fd-91da-6bd442fd2ca0/Marmarisboatcharter12-M https://www.wattpad.com/user/Marmarisboat https://www.goodreads.com/user/show/170488799-marmarisboatcharter12 https://gravatar.com/marmarisboatcharterc48f3b5147 https://tr.pinterest.com/besthairtransplant12/ https://www.mapleprimes.com/users/Marmarisboatcharter12 https://medium.com/@marmarisboatcharter https://www.ted.com/profiles/45229026 https://www.infragistics.com/community/members/478be3bd644f2f8748f42767e5de46f150b1c783?_ga=2.233792990.1346871303.1696583692-28579674.1696583692 https://www.metooo.io/u/651fd08c119b751a01cd570d https://app.roll20.net/users/12503055/marmarisboatcharter12-m https://list.ly/marmarisboatcharter/activity https://giphy.com/channel/Marmarisboatcharter12 https://www.tumblr.com/marmarisboatcharter12 https://www.creativelive.com/student/marmarisboatcharter12?via=accounts-freeform_2 https://gab.com/Marmarisboatcharter12 https://www.flickr.com/people/199297576@N06/ https://seedandspark.com/user/marmarisboatcharter12-01hc2f7yr1a08wjenetgcj2pw4 https://os.mbed.com/users/marmarisboatcharter1/ https://calis.delfi.lv/blogs/lietotajs/317879-marmarisboatcharter12/ https://notionpress.com/author/933488 https://my.desktopnexus.com/Marmarisboatcharter12/ https://guides.co/a/marmarisboatcharter12-marmarisboatcharter12/ https://www.bahamaslocal.com/userprofile/1/249936/Marmarisboatcharter12.html http://molbiol.ru/forums/index.php?showuser=1302210 https://www.credly.com/users/marmarisboatcharter12-marmarisboatcharter12/badges https://ko-fi.com/marmarisboatcharter12#paypalModal https://devpost.com/marmarisboatcharter?ref_content=user-portfolio&ref_feature=portfolio&ref_medium=global-nav https://www.redbubble.com/people/Marmarisboat/shop?asc=u https://hub.docker.com/u/marmarisboatcharter12 http://onlineboxing.net/jforum/user/editDone/254192.page https://micro.blog/Marmarisboat http://www.effecthub.com/user/3729468 http://hawkee.com/profile/5136037/ https://wefunder.com/marmarisboatcharter12 https://www.facer.io/user/4s2M4eDMJM https://mastodon.online/@Marmarisboatcharter12 https://sketchfab.com/Marmarisboatcharter12 https://profiles.wordpress.org/marmarisboatcharter12/ https://dzone.com/users/5002505/marmarisboatcharter12.html
Keywords
SRGB-based density analysis, Gaussian filter, Precision and F-measure
Introduction
In the field of medical science, the brain is identified as one of the significant organs in the human body which helps in processing and regulating complete body activities. Cancer is the deadliest disease that can occur in all organs of the human body by stimulating the growth of abnormal cells. Due to a variety of causes, 200 different types of cancer can affect the human body. Hence Cancer is considered as deadliest disease globally. As per the American Cancer Association statistics, nearly 80000 Americans were taken treatment for primary brain tumour stage and nearly 16000 Americans were dead from a brain tumour in the year 2019 [1].
Brain tumours are one of the most prevalent central nervous disorders. They can cause serious harm to human activities and occasionally even result in death. Among intracranial tumours, gliomas have the greatest death and morbidity rates. To identify brain tumours, several imaging techniques have been investigated with advanced technology. The most common techniques for diagnosing brain tumour disorders are Magnetic Resonance Imaging (MRI) and Computed Tomography (CT) scans. Structural images in three components are generated using the semi-imaging technique known as Magnetic Resonance Imaging (MRI). It is commonly used in the early identification, diagnosis, and follow-up of illnesses. It is founded on advanced edge technology that stimulates and detects abruptness in the rotational region of protons present in the water that composes tissues. Among them, MRI has emerged as the primary imaging method for the diagnosis and management of gliomas due to its benefits of smooth tissue contrast, imaging in any direction and non-invasive imaging
Related Works
Somasundaram and Gobinath presented are view analysis of deep cognition methods for tumour detection and segmentation [1]. This survey also studies current development in brain tumour analysis and categorization based on passing the MRI image patches into the deep networks. Lathera and Singh investigate the brain tumour detection and segmentation techniques proposed by many researchers in the past years [2]. This article also provides a consolidated explanation of partially and completely robotic brain tumour-finding techniques in tabular form. Yang and Song developed the U-net model for robotic brain tumour isolation [3]. The U-net architecture is constructed by using loss functions and an optimization algorithm for isolating the exact tumour region in brain MRI samples and implementing the semi-automatic brain tumour segmentation algorithm for identifying the accurate brain tumour space in MRI samples [4]. The result of this process is very close to the analysis of an expert radiologist. This technique is evaluated in terms of quantitative metrics.
Gobhinath et al. proposed a medical image-processing technique for tumour isolation in brain samples [5]. The high pass preprocessing concept is applied to reduce the unwanted elements and the median concept is applied to improve the superiority of the brain MRI sample. Akram and Usman implemented the CAD system for automatic brain tumour detection and isolation [6]. This diagnostic system helps to identify the tumour region with proper segmentation. Jemimma and Vetharaj implement the watershed conceptualizing Dynamic Angle Projection (DAPP) for brain tumour isolation. The Watershed concept effectively segments the tumour region by extracting texture features using Dynamic Angle Projection (DAPP) [7]. Convolution Neural Network (CNN) is applied to categorize the tumour region from the non-tumour region of the MRI image. Kumar et al. developed the fully Convolution Neural Network (CNN) for brain tumour isolation and classification [8]. The hierarchical approach and intra-growth hankie classification are applied for the exact categorization of tumour and non-tumour regions.
Wulandari et al. calculated the area of the brain tumour region by applying the watershed concept. The median concept is applied to improve the superiority of the MRI sample [9]. The thresholding technique is processed to calculate the accurate area of the brain tumour region introducing the two-step verification model based on the watershed algorithm for brain tumours segmentation [10]. This model isolated the tumour space in MRI samples and a matching algorithm is performed to compare the isolated area with the ground truth implemented in a semi-automated approach for brain tumour segmentation [11]. The data-driven model is implemented using the probabilistic method to isolate the brain tumour from images investigates the role of Convolution Neural Network (CNN) in brain tumour segmentation tasks and also investigates the future uses of convolution neural network by exploring radionics proposed by the tumour isolation network using A Res U-Net [12, 13]. Residual units are combined with U-net architecture to enhance the quality of the identification of brain tumour regions in MRI images giving the study the art techniques of pattern recognition for brain tumour segmentation in mp MRI samples [14]. This study also identifies the best learning model for brain tumour isolation in Brain Tumour Segmentation (BraTS) and challenges the proposed novel brain tumour isolation model [15]. This model is developed by combining the random field and multicascaded convolutional neural networks. The experimental results achieved competitive performance developed the combination of two segmentation techniques implemented for brain tumour isolation [16]. 3D Convolution Neural Network (CNN) and U-net architecture is combined for better and more accurate results. Ramya and Jayanthi implemented a semiautomated brain tumour segmentation model using graph cuts [17]. The kernel mapping technique is applied on graph cuts to partition the multi-regions for identifying the brain tumour region and developed an effective combined model for brain tumour segmentation [18]. SFCM spatial concept is applied to separate the tumour region from the non-tumour region.
Proposed Methodology
Brain tumour segmentation is a part of medical image processing. The automated or computerized brain tumour segmentation process helps medical practitioners to diagnose in the proper channel. Hence, this proposed method contains three stages to isolate the tumour space in brain MRI samples. In the first stage, the given input image is pre-processed into standard RGB colour space to narrow the intensity values. The Gaussian filter is applied on the grey level of the SRGB space image to normalize the tumour region in a uniform distribution. In the second stage, the possible tumour region is extracted by comparing the pre-processed outcome with the threshold value. Region properties such as density and area are employed to isolate the actual tumour space in the brain MRI samples. Finally, the morphological functions are applied to detect the exact tumour region by eliminating unwanted regions.
Preprocessing using colour space and gaussian filter
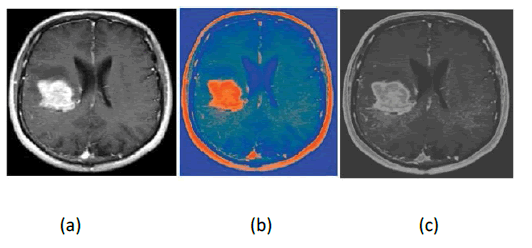
A colour value of an image furnishes more semantic information about objects and background scenes. Hence, image processing techniques entail the colour spaces along with certain properties for effective operations on images. Conversion from one colour space to another colour space needs philosophical knowledge of colour space characteristics and requirements of the particular application. Figure1 represents the outcome of SRGB and Gaussian filter.
Figure 1: Outcome of SRGB band Gaussian filter (a) Input (b) SRGB (c) Gaussian Filter
Colour space
In this proposed methodology, the present colour space shown in Figure 1(a) is converted into Standard RGB (SRGB) to represent the linear combination of colour values into the non- linear combination of colour values, which provides a more convenient way to categorize the required space in the brain MRI images. SRGB technique performs the non-linear function such as the gamma correction method on present colour space. This technique helps to distinguish the colour representation in brain MRI images. SRGB colour space is as shown in Figure 1(b) making the tumour region brighter as compared to the background region. The parameters used to transform the current colour space to SRGB space is equation 1, 2, and 3:
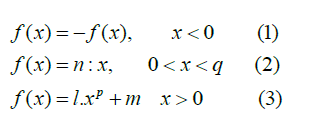
Where µ represents one of the R, G, or B colour values with the parameters:
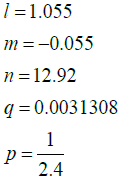
Gaussian filter
SRGB technique produces the sharpened high-frequency component tumour pixels, hence Gaussian filter is applied to smooth the pixels of the tumour region. Gaussian filter (equation 4) is a low pass filter, which reduces the noise by performing the blurriness on the image region as shown in Figure1(c). This technique is implemented with a symmetric kernel which is passed among all the pixels of the image to get the effective gap among the tumour and non-tumour pixels. Symmetric kernel changes the colour information due to the pixel that is the centre of the kernel having more weightage towards the final value.
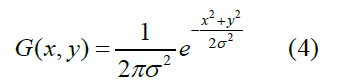
Possible tumour region extraction
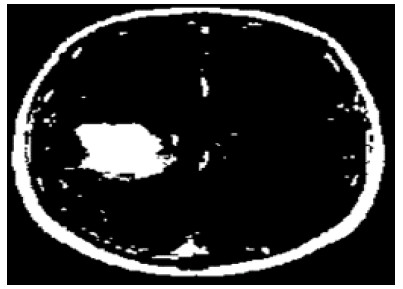
The thresholding technique is applied to extract the possible tumour region by comparing every pixel of the brain MRI sample with the computed value. Thresholding is a very popular technique to isolate the region of interest from the background region. Similarly, the threshold value is computed using equation 5. Later, each pixel value of Gaussian blurred outcome is compared with the computed threshold value. This separates all pixels of Gaussian blurred outcome into two sets such as pixels having intensity levels lower than the threshold and pixels having intensity levels greater than the threshold. Here, the proposed method considers the pixel values greater than the threshold value because the tumour region pixel intensity level is brighter than the background intensity level. Finally, its results with the binary image are shown in Figure 2.
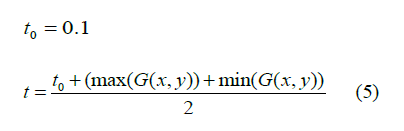
Figure 2: Extraction of the possible tumour region
Detection of the actual tumour region
The actual tumour region is extracted by analysing the region properties of the possible tumour region. Region-based properties such as area and solidity are studied to analyse the actual shape and position of the tumour space in brain MRI samples. The area region property identifies the actual number of intensities presenting the possible tumour samples. The convex area measures the shape factor by applying the convex polygon to a possible tumour region. Solidity computes the density of the tumour region by dividing the area over the convex area, which helps to retain the actual portion of the tumour region. Finally, the actual tumour region is extracted by intersecting the outcome of the solidity image with the possible tumour region as shown in The actual tumour region is extracted by analysing the region properties of the possible tumour region. Region-based properties such as area and solidity are studied to analyse the actual shape and position of the tumour space in brain MRI samples. The area region property identifies the actual number of intensities presenting the possible tumour samples. The convex area measures the shape factor by applying the convex polygon to a possible tumour region. Solidity computes the density of the tumour region by dividing the area over the convex area, which helps to retain the actual portion of the tumour region. Finally, the actual tumour region is extracted by intersecting the outcome of the solidity image with the possible tumour region as shown in Figure 3.
Figure 3: Outcome of the actual tumour region extraction
Post-processing for eliminating unwanted regions
After extracting the actual tumour region, there some unwanted regions were present in the tumour-detected image. Hence, morphological functions such as dilate, and filling operations are performed to eliminate the possible unwanted regions. Dilate operation combines all the disconnected tumour pixels. The filling operation loads the pixels into the holes created in the tumour region, which results in a complete region of the tumour area as shown in Figure 4.
Figure 4: Tumour region detected
Results
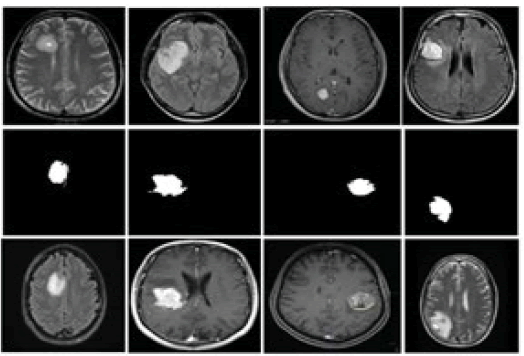
To assess the proposed methodology, an extensive experimental analysis is conducted on the Kaggle brain tumour MRI dataset. Which can be publicly downloaded in the linkage as follows in Figure 5. This dataset contains two different brain MRI classes Yes and No. Where Yes class represents the presence of a tumour region in brain MRI samples and No class depicts the absence of a tumour region in brain MRI samples. 253 brain MRI images were collected in this dataset with a standard size of 240 × 240 pixels. Out of 253 brains, MRI images 155 samples are Yes class with having a tumour region and 98 samples are No class with no brain tumour region. Sample outputs of the proposed approach are illustrated in Figure5.
Figure 5: Sample outputs of the proposed methodology
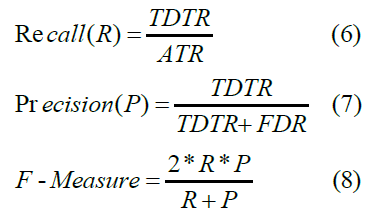
The evaluation metric of the proposed method is Recall (R) (Equation 6): Truly Detected Tumour Region (TDTR) over the Actual Tumour Region (ATR). Precision (P) (Equation 7): Truly Detected Tumour Region (TDTR) over the Truly Detected Tumour Region (TDTR) and Falsely Detected Region (FDR). F-Measure (Equation 8): Product of 2, Recall and Precision over the addition of Recall and Precision.
The analysis result of the proposed methodology is represented in Table 1 concerning both classes. SRGB colour space effectively increases the semantic gap between the tumour region and nonregion. Morphological functions also eliminate all possible false positives to increase the result precision. In Table1, separate result analysis has been done for both yes and no classes and also aver-aged results have also been calculated to represent the overall performance of the proposed approach.
Tab 1. Evaluation of proposed methodology
| Classes | Precision | Recall | F-Measure |
|---|---|---|---|
| Yes | 90.14 | 83.11 | 86.48 |
| No | 89.53 | 78.57 | 83.69 |
| Average | 89.83 | 80.84 | 85.08 |
The comparative analysis of the presented approach is also done in Table 2 with previous state-of-the-art techniques. The obtained results are competitive concerning the recall and outperform concerning precision and F-measure.
Tab. 2. Comparative analysis of Proposed Methodology
| Methods | Precision | Recall | F-Measure |
|---|---|---|---|
| K-Means[19] | 0.83 | 0.64 | 0.72 |
| K-Means with PSO[19] | 0.86 | 0.82 | 0.84 |
| DWT+PCA+SVM (Linear) [20] | 0.4 | 1 | 0.57 |
| Proposed Methodology | 89.83 | 80.84 | 85.08 |
Conclusion
The automated brain tumour segmentation techniques are more helpful to medical practitioners to diagnose the patient in the proper channel. Hence, the currently proposed methodology implements the SRGB colour space with Gaussian filter. SRGB effectively distinguishes the tumour intensity values from the non-tumour intensity values. Gaussian filter is helpful to smooth the pixels of the tumour region. Later threshold value completely differentiates the tumour pixels from the nontumour pixels and it is considered a possible tumour region. Area and density of region properties effectively identify the accurate tumour region. Morphological functions are helpful to eliminate the possible false positives in the outcome. In future, post-processing techniques will be studied to extract the actual region tumour region from complex MRI images.
References
- Somasundaram S, Gobinath R. Current trends on deep learning models for brain tumour segmentation and detectionâ??a review. IEEE. 2019: 217-221.
- Lather M, Singh P. Investigating brain tumour segmentation and detection techniques. Procedia Comput Sci. 2020; 167:121-30.
- Yang T, Song J. An automatic brain tumour image segmentation method based on the U-Net. IEEE. 2018;1600-1604
- Ganasala P, Kommana DS, Gurrapu B. Semiautomatic and automatic brain tumour segmentation methods: performance comparison. IEEE. 2020;43-46.
- Gobhinath S, Anandkumar S, Dhayalan R, Ezhilbharathi P, Haridharan R. Human Brain Tumour Detection and Classification by Medical Image Processing. IEEE. 2021;561-564
- Akram MU, Usman A. Computer aided system for brain tumour detection and segmentation. IEEE. 2011;299-302.
- Jemimma TA, Vetharaj YJ. Watershed algorithm based DAPP features for brain tumour segmentation and classification. IEEE. 2018;155-158.
- Kumar S, Negi A, Singh JN, Gaurav A. Brain tumour segmentation and classification using MRI images via fully convolution neural networks. IEEE.2018;1178-1181.
- Wulandari A, Sigit R, Bachtiar MM. Brain tumour segmentation to calculate percentage tumour using MRI. IEEE. 2018;292-296.
- Hasan SM, Ahmad M. Two-step verification of brain tumour segmentation using watershed-matching algorithm. Brain Inform. 2018; 5:1-1.
- Solomon J, Butman JA, Sood A. Data driven brain tumour segmentation in mri using probabilistic reasoning over space and time. Springer.2004;301-309.
- Bhandari A, Koppen J, Agzarian M. Convolutional neural networks for brain tumour segmentation. Insights Imaging. 2020; 11:1-9.
- Zhang J, Lv X, Zhang H, Liu B. AResU-Net: Attention residual U-Net for brain tumour segmentation. Symmetry. 2020; 12:721.
- Bakas S, Reyes M, Jakab A, Bauer S, Rempfler M, et al. Identifying the best machine learning algorithms for brain tumour segmentation, progression assessment, and overall survival prediction in the BRATS challenge. ArXiv prepr. 2018.
- Hu K, Gan Q, Zhang Y, Deng S, Xiao F, et al. Brain tumour segmentation using multi-cascaded convolutional neural networks and conditional random field. IEEE Access. 2019; 7:92615-92629.
- Ali M, Gilani SO, Waris A, Zafar K, Jamil M. Brain tumour image segmentation using deep networks. IEEE Access. 2020; 8:153589-153598.
- Ramya R, Jayanthi KB. Multiregion Image Segmentation by Graph Cuts for Brain Tumour Segmentation. Springer. 2012;329-332.
- Singh N, Das S, Veeramuthu A. An efficient combined approach for medical brain tumour segmentation. IEEE. 2017; 1325-1329.
- Kapoor A, Agarwal R. Enhanced Brain Tumour MRI Segmenta tion using K-means with machine learning based PSO and Firefly Algorithm. EAI Endorsed Trans. Pervasive Health Technol. 2021;7.
- Ejaz K, Rahim MS, Rehman A, Chaudhry H, Saba T, et al. Segmentation method for pathological brain tumour and accurate detection using MRI. Int J Adv Comput Sci Appl. 2018;9:394-401.